Difference between revisions of "Constructing a Pathway of Intermediate States"
m (moved Intermediate States to Constructing a Pathway of Intermediate States: Not clear) |
|
(No difference)
| |
Revision as of 19:56, 8 October 2012
| Free Energy Fundamentals |
|---|
 |
|
Methods of Free Energy Simulations
|
| Free Energy How-to's |
|---|
 |
Because the phase space between two target states of interest can be near zero, doing free energy calculations for the two states alone will often have very large errors. When one considers the large number of potential molecules that could be considered interesting, this problem becomes even more pronounced. Because free energy is a state function, we can construct a thermodynamic path that takes us through a set of states that improves phase space overlap between states. To put this mathematically, we can improve our results by constructing high phase space overlap intermediates and calculating our free energy difference by
- [math]\displaystyle{ \Delta A_{1,K} = \sum_{i=1}^{K-1} \Delta A_{i,i+1} }[/math].
One advantage available to computational free energy calculations is the ability to simulate unphysical states. By this, we mean that our intermediate states do not have to be observable experimentally. This is a fact that most if not all computational methods out there take advantage of. Again, because free energy is a state function, we simply choose the path of greatest convenience and carry out the calculation across this path. However, choosing the "correct" or even a "good" path is a very challenging action and is one of the most difficult tasks in the entire field of free energy calculations.
Because our free energy calculations frequently involve transforming one kind of atom at a given site to another kind, the transformations are often referred to as "alchemical."
Linear Alchemical Potential
To be more clear about what it means to define an "alchemical path," one should think of it as defining the thermodynamic path where we modify, remove, or add various forces on an atom. Take for instance estimating the free energy difference between a Lennard-Jones fluid and a Stockmayer fluid, our thermodynamic path would calculate the work and free energy required to switch on the dipole interactions at a rate that maximizes phase space overlap and efficiency.[1] Because the atoms changed their interactions with the surroundings without being removed or added from the system, we have directly modified the atoms to create our alchemical path.
Most alchemical transformations can be defined by alchemically scaling the potential in some manner. The simplest of these is the linear transformation, which says that the net potential energy, [math]\displaystyle{ U(\lambda,\vec{q}) }[/math], is the sum of the alchemically modified two end states' potentials, [math]\displaystyle{ U_0 }[/math] and [math]\displaystyle{ U_1 }[/math], plus the parts of the potential that are unaffected by the alchemical transformation, [math]\displaystyle{ U_{\mathrm{unaffected}} }[/math]; or
- [math]\displaystyle{ U(\lambda,\vec{q}) = (1-\lambda) U_0(\vec{q}) + \lambda U_1(\vec{q}) + U_{\mathrm{unaffected}}(\vec{q}) }[/math]
where the alchemical variable linearly modifies the confrontational information from each state of interest.
Problems with Linear Transformations
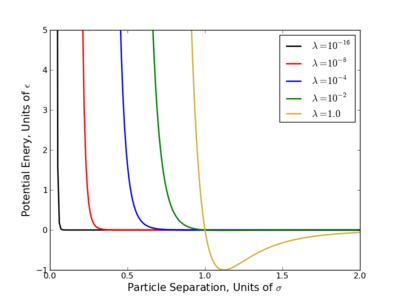
The largest problem with linear scaling is that it does not provide equal phase space overlap between states. This has been a long acknowledged problem with several easy to see reasons why.
- Consider the OPLS-AA force field, at [math]\displaystyle{ \lambda=0.1 }[/math], the excluded volume of a methane sphere is still 60-70%.
- Most potential energy functions have an infinite potential (singularity) at [math]\displaystyle{ r=0 }[/math]. This is usually brought on by the [math]\displaystyle{ r^{-12} }[/math] term that appears in Lennard-Jones based potentials and is present at all nonzero values of [math]\displaystyle{ \lambda }[/math]. Since most the free energy techniques require some numeric methods, evaluating this singularity diverges the calculations.
This singularity causes the most problems with TI since that method requires evaluation [math]\displaystyle{ \frac{dU}{d\lambda} }[/math] numerically, which diverges at the singularity. Several workaround for this have been suggested where one of the simplest is do raise the [math]\displaystyle{ \lambda }[/math] terms to a power greater than or equal to 4 like so
- [math]\displaystyle{ \displaystyle U(\lambda) = (1-\lambda)^4 U_0 + \lambda^4 U_1 }[/math]
These methods do converge [math]\displaystyle{ \left\langle\frac{dU}{d\lambda} \right\rangle }[/math] however very slowly with number of samples and still can cause numeric instabilities[3][4]
Other methods have been suggested such as shrinking the core of the tom, but this also causes instabilities[3][5][6] and are not practical for systems with many bonds.
Soft Core Potentials
A standard method has been developed to alchemically change molecules to get around the numeric instabilities called a "soft core potential."[2][7] In these potentials, the configuration variable, [math]\displaystyle{ r }[/math], is now coupled to the alchemical variable, [math]\displaystyle{ \lambda }[/math], smoothing out the singularity and looks like
- [math]\displaystyle{ U(\lambda,r) = 4\epsilon\lambda^n \left[\left(\alpha(1-\lambda)^m + \left(\frac{r}{\sigma}\right)^6\right)^{-2} + \left(\alpha(1-\lambda)^m + \left(\frac{r}{\sigma}\right)^6\right)^{-1}\right] }[/math]
where [math]\displaystyle{ \alpha }[/math] is a constant (usually 0.5) and, [math]\displaystyle{ m }[/math] and [math]\displaystyle{ n }[/math] are positive integers with the original choice being [math]\displaystyle{ n=4 }[/math] and [math]\displaystyle{ m=2 }[/math]. Recent improvements have shown that keeping [math]\displaystyle{ \alpha=0.5 }[/math] and setting [math]\displaystyle{ m=n=1 }[/math] provides significant improvement to the variance[3][4][8] and research into further improving this is still going on.[9]
This potential is now considered the standard for avoiding endpont errors, and some version of this is common in many software packages that support free energy calculations As a naming convention, when "soft core" is referenced, it refers to the this potential where as "linear" refers to the linear alchemical potential where the exponent is 1 (i.e. [math]\displaystyle{ \lambda }[/math] not [math]\displaystyle{ \lambda^4 }[/math]).
Rules of Thumb for Intermediate States
This section is just for the short version of the discussions found elsewhere(mostly below) on the site. These rules are not the end-all set and you should be familiar with why each one is suggested before just accepting them.
- Bonded terms can be modified/turned off linearly. This includes angle or bond force constants as well as unconstrained bond distances.
- Constrained bonds should not change length. There are free energy changes that cannot be ignored affiliated with this action.[10]
- Maximize similarity between states by removing/decoupling as few atoms as possible.
- Do not open and close rings. This supersedes the previous rule.
- Statistical uncertainty between any neighboring states should be equal. Rather challenging to do, but it has been proven to have the lowest variance path if you can pull it off.[11]
- Deleting or adding atoms should always be done with a soft core potential.
- Changes in parameters can be done linearly.
- All charge on atoms must be turned off prior to atomic repulsion. Otherwise you can get an infinite attractive potential and crash your simulation.
- Similarly for only changes in terms, its generally more efficient to change electrostatic terms separate from Lennard-Jones terms.
- More states is better than fewer. Variance shrinks rapidly with number of states. You want the difference between intermediaries to be between 2-3 [math]\displaystyle{ k_BT }[/math]. Obviously you will be limited on CPU power, but fewer states also leads to more samples begin required from each state, so take this into account when deciding number of states as well.
- Shape of the variance does not significantly change with number of atoms, only magnitude.[12] More intermediates will still be required for a large number of atoms to reduce statistical noise.
- Charge should be maintained across all [math]\displaystyle{ \lambda }[/math]. Simply having charged molecules is fine, but the net of the system should remain constant. If you must change the net charge, there are complicated ways to do so.[13][14]
- Short prototype simulations are recommended. Even as short as 100 ps, the prototypes can provide rough magnitude of variance estimates, although will likely under-predict the free energy as many configurations remain unsampled.
Constructing Intermediate States
This section outlines some of the options one has when constructing their thermodynamic path through the intermediary states. Each method has its own strengths and weakness and should be carefully considered before implementing. Although there is no absolute best way to do this, there are a number of best practices we strongly consider following, especially for those starting out in free energy calculations. In order to fully understand some of the choices in this section relative to the grand scheme of things, please read the thermodynamic cycle page.
Intermolecular Forces
When designing intermediate states, one must decide early on which forces to change, and how to change them. For intermolecular forces, both the electrostatic and Lennard-Jones interactions must be turned off, however, one should not change all the forces at the same time. If one were to turn off the repulsive Lennard-Jones components first, unlike charges on atoms would be infinitely attracted to each other and cause the simulation to crash. As such, one should perform one of the following sequences when decoupling atoms and/or molecules:
Decoupling Scheme A
- Disable all electrostatics linearly
- Disable Lennard-Jones terms by soft core
Decoupling Scheme B
- Disable electrostatics AND dispersion (attractive) terms together linearly
- Disable the repulsive components by soft core
Either method is rather efficient[15][4] and can be followed in reverse for appearing molecules. Although all the terms could be turned off by soft core at the same time to avoid negative infinities,[6][16] parametrization is a pain and hard to generalize.[17]
For those just starting, we highly recommend Decoupling Scheme A.
Topologies
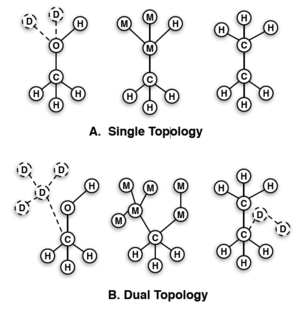
After the intermolecular forces are decided, you will then need to decide the overall method in which you will modify your molecules. There are two main approaches known as the single topology approach and the dual topology approach; examples of this can be seen in the figure to the side which will be referenced accordingly. Single topology (A, upper) has only one site for any location shared between the end states, and then "dummy" atoms to make up for any unique sites. During the transformation, the dummy atoms are transformed into fully interacting atoms, and the shared site atom is transformed directly to a new atom. Dual topology (B, lower) is different in that shared sites between states do not share atoms. No atom changes its type, just the interactions with the surrounding system.
The dual topology does require more dummies, which means more CPU power and needed intermediates, however, it has a very strong advantage in that the dummies can simultaneously explore more congregational space while decoupled; this perk is amplified when simulations are run with Hamiltonian exchange or expanded ensemble. One may notice that, even though the dummies have nonbonded interactions, they are still "bound" to other atoms which make the end states have slightly nonphysical character. These interactions though can be accounted for by simulating in both vacuum and in molecular surrounds. With fixed bond lengths in the rigid rotor approximation, the effect of these dummies on the free energy is negated.[19] If the bonds are not constrained, there is a difference, but not enough to be of concern (>0.01 kcal/mol).
In either topology, it may also be necessary to modify the bonded terms (especially for angle and dihedral terms). Because of the time scale of these motions, these terms converge quickly leaving one with small variances but large energy changes. For simple bonded parameters like harmonic spring constants and equilibrium bond lengths, linear changes are perfectly acceptable. Constrained bonds are challanging to compute as correction terms are needed from the complete lack of phase space overlap[20]
Either topological approach will provide the same results and which one is used will often depend on the simulation package/code.
Alchemically Transforming Rings
Rings in free energy calculations are rather challenging as they prove the exception to the rule of "disappear as few atoms as possible." The act of opening or closing a ring alchemically involves drastic changes to the phase space overlap which should be avoided if at all possible. As such, rings should be disappeared entirely and the new molecule appeared if you find the need to actually break the ring, and in reverse for appearing the ring. This has been shown to be straightforward with the soft core potentials and is still accurate despite the large number of transformations.[8][12][21][22][23]
Pulling Methods
A rather different approach to the intermediate states is to set up a system where the ligand is physically pulled away from the protein. The two end states are then where the ligand is in its binding pocket (or nearby at least), and then another where the ligand is far enough away to be considered not interacting with the protein; the free energy in this system can be seen as free energy of binding. Carrying these out can be done with either nonequilibrium methods,[24] or by setting up an umbrella simulation with different overlapping harmonic oscillators at increasing distances from the binding pocket, then calculating the PMF.[25][26][27]
Although this may seem simple enough, there are a number of problems with pulling methods and so this method is not recommended for those just starting in free energy methods. That said, this method is still valid and has even been suggested to be very efficient for highly charged ligands.[26]
Challenges for the pulling methods
- Direct exit path for the ligand from the binding pocket may be difficult to identify. This results in large statistical error.
- Large box sizes are needed for this method to approximate the ligand and protein not interacting.
- If smaller boxes are required (limited computational resources), mean-field or analytical approximations must be made to estimate the "infinite distance away" ligand free energy.
References
- ↑ Frenkel, D., & Smit, B. (2002). Understanding Molecular Simulation (2nd ed., p. 638). San Diego: Academic Press.
- ↑ 2.0 2.1 Beutler, T., Mark, A., & Schaik, R. van. (1994). Avoiding singularities and numerical instabilities in free energy calculations based on molecular simulations. Chemical Physics Letters, 222(June), 529–539.
- ↑ 3.0 3.1 3.2 Pitera, J. W., and van Gunsteren, W. F. (2002) A comparison of non-bonded scaling approaches for free energy calculations. Mol. Simulat. 28, 45–65.
- ↑ 4.0 4.1 4.2 Steinbrecher, T., Mobley, D. L., and Case, D. A. (2007) Nonlinear scaling schemes for Lennard-Jones interactions in free energy calculations. J. Chem. Phys. 127, 214108.
- ↑ Pearlman, D. A., and Connelly, P. R. (1995) Determination of the differential effects of hydrogen bonding and water release on the binding of FK506 to native and TYR82 → PHE82 FKBP-12 proteins using free energy simulations. J. Mol. Biol. 248, 696–717.
- ↑ 6.0 6.1 Wang, L., and Hermans, J. (1994) Change of bond length in free-energy simulations: Algorithmic improvements, but when is it necessary? J. Chem. Phys. 100, 9129–9139.
- ↑ Zacharias, M., Straatsma, T. P., and McCammon, J. A. (1994) Separation-shifted scaling, a new scaling method for Lennard-Jones interactions in thermodynamic integration. J. Phys. Chem. 100, 9025–9031.
- ↑ 8.0 8.1 Shirts, M. R., and Pande, V. S. (2005) Solvation free energies of amino acid side chains for common molecular mechanics water models. J. Chem. Phys. 122, 134508.
- ↑ Pham, T. T., and Shirts, M. R. (2011) Identifying Low Variance Pathways for Free Energy Calculations of Molecular Transformations in Solution Phase. J. Chem. Phys. 135, 034114.
- ↑ Boresch, S., and Karplus, M. (1996) The Jacobian factor in free energy simulations. J. Chem. Phys. 105, 5145–5154.
- ↑ Shenfeld, D. K., Xu, H., Eastwood, M. P., Dror, R. O., and Shaw, D. E. (2009) Minimizing thermodynamic length to select intermediate states for free-energy calculations and replica-exchange simulations. Phys. Rev. E 80, 046705.
- ↑ 12.0 12.1 Shirts, M. R., Pitera, J. W., Swope, W. C., and Pande, V. S. (2003) Extremely precise free energy calculations of amino acid side chain analogs: Comparison of common molecular mechanics force fields for proteins. J. Chem. Phys. 119, 5740–5761.
- ↑ Kastenholz, M. A., and Hünenberger, P. H. (2006) Computation of methodology-independent ionic solvation free energies from molecular simulations. I. The electrostatic potential in molecular liquids. J. Chem. Phys. 124, 124106.
- ↑ Kastenholz, M. A., and Hünenberger, P. H. (2006) Computation of methodology-independent ionic solvation free energies from molecular simulations. II. The hydration free energy of the sodium cation. J. Chem. Phys. 124, 224501.
- ↑ Wang, J., Deng, Y., and Roux, B. (2006) Absolute Binding Free Energy Calculations Using Molecular Dynamics Simulations with Restraining Potentials. Biophys. J. 91, 2798–2814.
- ↑ Blondel, A. (2004) Ensemble variance in free energy calculations by thermodynamic integration: theory, optimal Alchemical path, and practical solutions. J. Comput. Chem. 25, 985–993.
- ↑ Steinbrecher, T., Joung, I., & Case, D. a. (2011). Soft-core potentials in thermodynamic integration: comparing one- and two-step transformations. J. of Comp. Chem., 32(15), 3253–63. doi:10.1002/jcc.21909
- ↑ Baron, R. (Ed.). (2012). Computational Drug Discovery and Design (p. 628). New York: Humana Press. doi:10.1007/978-1-61779-465-0
- ↑ Boresch, S., and Karplus, M. (1999) The Role of Bonded Terms in Free Energy Simulations. 2. Calculation of Their Influence on Free Energy differences of Solvation. J. Phys. Chem. A 103, 119–136.
- ↑ Boresch, S., Tettinger, F., Leitgeb, M., and Karplus, M. (2003) Absolute binding free energies: A quantitative approach for their calculation. J. Phys. Chem. A 107, 9535–9551.
- ↑ Mobley, D. L., Dumont, È., Chodera, J. D., and Dill, K. A. (2007) Comparison of charge models for fixed-charge force fields: Small-molecule hydration free energies in explicit solvent. J. Phys. Chem. B 111, 2242–2254.
- ↑ Nicholls, A., Mobley, D. L., Guthrie, P. J., Chodera, J. D., and Pande, V. S. (2008) Predicting Small-Molecule Solvation Free Energies: An Informal Blind Test for Computational Chemistry. J. Med. Chem. 51, 769–779.
- ↑ Mobley, D. L., Bayly, C. I., Cooper, M. D., and Dill, K. A. (2009) Predictions of Hydration Free Energies from All-Atom Molecular Dynamics Simulations˘a. J. Phys. Chem. B 113, 4533–4537.
- ↑ Ytreberg, F. (2009) Absolute FKBP binding affinities obtained via nonequilibrium unbinding simulations. J. Chem. Phys. 130, 164906.
- ↑ Lee, M. S., and Olson, M. A. (2006) Calculation of Absolute Protein-Ligand Binding Affinity Using Path and Endpoint Approaches. Biophys. J. 90, 864–877.
- ↑ 26.0 26.1 Woo, H.-J., and Roux, B. (2005) Calculation of absolute protein-ligand binding free energy from computer simulation. Proc. Natl. Acad. Sci. 102, 6825–6830.
- ↑ Gan, W., and Roux, B. (2008) Binding specificity of SH2 domains: Insight from free energy simulations. Proteins 74, 996–1007.