File list
Jump to navigation
Jump to search
This special page shows all uploaded files.
| Date | Name | Thumbnail | Size | User | Description | Versions |
|---|---|---|---|---|---|---|
| 17:03, 22 December 2013 | Automated-equilibration-detection.png (file) |  |
38 KB | John Chodera | Top: Statistical inefficiency <math>g</math>. Bottom: Number of effectively uncorrelated samples <math>N_{eff}</math>. | 1 |
| 20:50, 22 December 2013 | Replica-round-trips.png (file) | 55 KB | John Chodera | Illustration of three replica trajectories in thermodynamic state index (y-axis) plotted as a function of simulation time (x-axis) for a Hamiltonian exchange absolute alchemical free energy calculation for p-xylene binding to T4 lysozyme L99A. | 1 | |
| 21:04, 22 December 2013 | Gelman-rubin.png (file) | 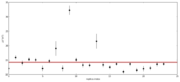 |
18 KB | John Chodera | 1 | |
| 21:05, 22 December 2013 | Gelman-rubin-z-score.png (file) | 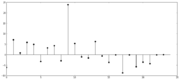 |
15 KB | John Chodera | 1 | |
| 21:35, 22 December 2013 | Hindered-sidechain-torsion.png (file) | 19 KB | John Chodera | Replica trajectories for the <math>\chi_1</math> angle of T4 lysozyme L99A binding to p-xylene in a Hamiltonian exchange simulation of an absolute alchemical free energy calculation in implicit solvent. | 1 | |
| 19:41, 7 March 2014 | Minimal.tar.gz (file) | 14.75 MB | David Mobley | T4 L99A/M102Q minimal set of topology/coordinate files. | 1 | |
| 17:08, 11 March 2014 | CCP.zip (file) | 29.48 MB | David Mobley | CCP model binding site minimal set of inputs | 1 | |
| 06:06, 14 March 2014 | Absolute n-phenylglicinonitrile T4 lysozyme cycle.pdf (file) |  |
612 KB | Matteo | .png version | 2 |
| 06:24, 14 March 2014 | T4+nphenylglicinonitrile ligand.results.doc (file) | 3 KB | Matteo | Summary of results for the ligand in water calculations in the tutorial GROMACS 4.6. n-phenylglicinonitrile with T4 lysozyme | 1 | |
| 11:09, 18 May 2014 | FKBP desmond.tgz (file) | 11.08 MB | Michael Shirts | Desmond compatible files for running FKBP. | 1 | |
| 10:19, 25 May 2014 | Shirts Workshop dG Methods 2014.pdf (file) | 14.79 MB | Michael Shirts | Michael Shirts's talk from Wednesday, May 21st, 2014 at the Workshop for Free Energy Methods in Drug Design | 1 | |
| 20:01, 29 May 2014 | Boston FEC 051914 06 RonLevy.pptx (file) | 4.91 MB | Michael Shirts | Ron Levy Presentation, 2014 Workshop on Free Energy Methods in Drug Design | 1 | |
| 22:17, 29 May 2014 | GabrielRocklin2014-PeriodicityArtifacts.pdf (file) | 680 KB | Grocklin | 2014 Free Energy Workshop slides | 1 | |
| 10:08, 13 June 2014 | Neale Vertex 2014.pptx (file) | 11.51 MB | Cneale | 1 | ||
| 10:13, 14 June 2014 | InputFiles T4 lysozyme+nphenylglicinonitrile.tar.gz (file) | 976 KB | Matteo | 2 | ||
| 13:04, 16 June 2014 | Protein-Ligand Binding Free Energies for the Masses.pdf (file) | 12.45 MB | Daveminh | 1 | ||
| 15:42, 16 June 2014 | T4+nphenylglicinonitrile complex.results.doc (file) | 5 KB | Matteo | 2 | ||
| 19:30, 18 December 2014 | 2014 Vertex Mobley.pdf (file) | 10.44 MB | Lnaden | David Mobley's slides. Recovered from before the pre-spam backup restore. | 1 | |
| 18:59, 10 March 2016 | Holden.png (file) |  |
27 KB | Mjschnie | 1 | |
| 19:01, 10 March 2016 | Laufer.png (file) | 7 KB | Mjschnie | 1 | ||
| 19:07, 10 March 2016 | Pfizer.png (file) |  |
19 KB | Mjschnie | 1 | |
| 19:07, 10 March 2016 | OpenEye.png (file) |  |
8 KB | Mjschnie | 1 | |
| 19:08, 10 March 2016 | Astex.png (file) |  |
6 KB | Mjschnie | 1 | |
| 11:48, 15 May 2016 | Merck Serono postdoctoral researcher for predictive models in drug design.pdf (file) | 517 KB | John Chodera | Merck Serono postdoctoral researcher for predictive models in drug design - 2016-06-15 | 1 | |
| 11:50, 15 May 2016 | Merck Serono postdoctoral researcher in computational chemistry and molecular dynamics.pdf (file) | 515 KB | John Chodera | Merck Serono postdoctoral researcher in computational chemistry and molecular dynamics - 2016-05-15 | 1 | |
| 13:09, 15 May 2016 | VertexComputationalChemist JobFlyer.pdf (file) | 242 KB | John Chodera | Vertex - Research Scientist I, Computational Chemist/Modeling and Informatics (posted 2016-05-15) | 1 | |
| 14:16, 20 May 2016 | Sampling Discussion.pdf (file) | 36 KB | Michael Shirts | Summary of the sampling discussion on Tuesday afternoon | 1 | |
| 15:11, 20 May 2016 | YANK-tutorial-2016-05-20.pdf (file) | 424 KB | John Chodera | YANK tutorial slides presented at 2016 Markov state models and kinetics in drug discovery workshop at Novartis. | 1 | |
| 22:45, 26 May 2016 | 2016 Vertex.pdf (file) | 26.56 MB | David Mobley | Mobley's 2016 Vertex talk | 1 | |
| 19:19, 8 June 2016 | Vertex-workshop-Baofeng-Zhang-51316.pdf (file) | 12.49 MB | David Mobley | 1 | ||
| 19:20, 8 June 2016 | Vertex FreeEnergyWorkshop2016 AV.pdf (file) | 708 KB | David Mobley | 1 | ||
| 19:21, 8 June 2016 | Poster LaurentiuSpiridon.pdf (file) | 1.42 MB | David Mobley | 1 | ||
| 19:13, 28 September 2016 | Vertex talk 5 15 2016 clean3.pdf (file) | 2.81 MB | David Mobley | Robert Abel partial slides from Vertex workshop | 1 | |
| 05:26, 20 February 2017 | Therm cycle.pdf (file) |  |
1.02 MB | Matteo | png | 2 |
| 05:39, 20 February 2017 | Results ABFE GMX2016.zip (file) | 209 KB | Matteo | Sample results for the tutorial "Absolute Binding Free Energy - Gromacs 2016" | 1 | |
| 05:43, 20 February 2017 | Results HREX ABFE GMX2016.zip (file) | 113 KB | Matteo | Example alchemical_analysis.py results (sims with HREX) for the tutorial "Absolute Binding Free Energy - Gromacs 2016" | 1 | |
| 05:50, 20 February 2017 | Cp xvg.sh.zip (file) | 301 bytes | Matteo | simple example script to add to the tutorial "Absolute Free Energy Calculations - Gromacs 2016" | 1 | |
| 04:13, 23 February 2017 | InputFiles ABFE GMX2016.zip (file) | 922 KB | Matteo | 3 | ||
| 15:39, 12 September 2017 | Novartis-logo.png (file) | 25 KB | Mjschnie | 2 | ||
| 10:04, 13 September 2017 | SiliconTx.png (file) | 42 KB | Mjschnie | 4 | ||
| 16:52, 7 February 2018 | Acellera 500px.png (file) | 21 KB | Mjschnie | Acellera | 1 | |
| 17:01, 7 February 2018 | Cresset Software Logo 500px.jpg (file) |  |
68 KB | Mjschnie | 2 | |
| 17:02, 7 February 2018 | Entasis Therapeutics 500px.jpg (file) |  |
60 KB | Mjschnie | 1 | |
| 17:09, 7 February 2018 | Vertex.png (file) |  |
32 KB | Mjschnie | 3 | |
| 17:11, 7 February 2018 | Vertex 500px.png (file) |  |
32 KB | Mjschnie | 1 | |
| 17:53, 7 February 2018 | Schrodinger 500px.png (file) | 25 KB | Mjschnie | 1 | ||
| 10:52, 19 February 2018 | Biki 500px.jpg (file) |  |
572 KB | Mjschnie | 1 | |
| 11:54, 19 February 2018 | AstraZeneca 500px.png (file) | 32 KB | Mjschnie | 1 | ||
| 14:37, 29 March 2018 | Openeye-logo-large-500px.jpg (file) |  |
45 KB | Mjschnie | 1 | |
| 16:18, 3 April 2018 | Genetech-500px.jpg (file) |  |
22 KB | Mjschnie | 1 |