File list
Jump to navigation
Jump to search
This special page shows all uploaded files.
| Date | Name | Thumbnail | Size | User | Description | Versions |
|---|---|---|---|---|---|---|
| 18:59, 10 March 2016 | Holden.png (file) |  |
27 KB | Mjschnie | 1 | |
| 19:30, 18 December 2014 | 2014 Vertex Mobley.pdf (file) | 10.44 MB | Lnaden | David Mobley's slides. Recovered from before the pre-spam backup restore. | 1 | |
| 15:42, 16 June 2014 | T4+nphenylglicinonitrile complex.results.doc (file) | 5 KB | Matteo | 2 | ||
| 13:04, 16 June 2014 | Protein-Ligand Binding Free Energies for the Masses.pdf (file) | 12.45 MB | Daveminh | 1 | ||
| 10:13, 14 June 2014 | InputFiles T4 lysozyme+nphenylglicinonitrile.tar.gz (file) | 976 KB | Matteo | 2 | ||
| 10:08, 13 June 2014 | Neale Vertex 2014.pptx (file) | 11.51 MB | Cneale | 1 | ||
| 22:17, 29 May 2014 | GabrielRocklin2014-PeriodicityArtifacts.pdf (file) | 680 KB | Grocklin | 2014 Free Energy Workshop slides | 1 | |
| 20:01, 29 May 2014 | Boston FEC 051914 06 RonLevy.pptx (file) | 4.91 MB | Michael Shirts | Ron Levy Presentation, 2014 Workshop on Free Energy Methods in Drug Design | 1 | |
| 10:19, 25 May 2014 | Shirts Workshop dG Methods 2014.pdf (file) | 14.79 MB | Michael Shirts | Michael Shirts's talk from Wednesday, May 21st, 2014 at the Workshop for Free Energy Methods in Drug Design | 1 | |
| 11:09, 18 May 2014 | FKBP desmond.tgz (file) | 11.08 MB | Michael Shirts | Desmond compatible files for running FKBP. | 1 | |
| 06:24, 14 March 2014 | T4+nphenylglicinonitrile ligand.results.doc (file) | 3 KB | Matteo | Summary of results for the ligand in water calculations in the tutorial GROMACS 4.6. n-phenylglicinonitrile with T4 lysozyme | 1 | |
| 06:06, 14 March 2014 | Absolute n-phenylglicinonitrile T4 lysozyme cycle.pdf (file) |  |
612 KB | Matteo | .png version | 2 |
| 17:08, 11 March 2014 | CCP.zip (file) | 29.48 MB | David Mobley | CCP model binding site minimal set of inputs | 1 | |
| 19:41, 7 March 2014 | Minimal.tar.gz (file) | 14.75 MB | David Mobley | T4 L99A/M102Q minimal set of topology/coordinate files. | 1 | |
| 21:35, 22 December 2013 | Hindered-sidechain-torsion.png (file) | 19 KB | John Chodera | Replica trajectories for the <math>\chi_1</math> angle of T4 lysozyme L99A binding to p-xylene in a Hamiltonian exchange simulation of an absolute alchemical free energy calculation in implicit solvent. | 1 | |
| 21:05, 22 December 2013 | Gelman-rubin-z-score.png (file) | 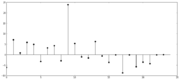 |
15 KB | John Chodera | 1 | |
| 21:04, 22 December 2013 | Gelman-rubin.png (file) | 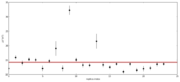 |
18 KB | John Chodera | 1 | |
| 20:50, 22 December 2013 | Replica-round-trips.png (file) | 55 KB | John Chodera | Illustration of three replica trajectories in thermodynamic state index (y-axis) plotted as a function of simulation time (x-axis) for a Hamiltonian exchange absolute alchemical free energy calculation for p-xylene binding to T4 lysozyme L99A. | 1 | |
| 17:03, 22 December 2013 | Automated-equilibration-detection.png (file) |  |
38 KB | John Chodera | Top: Statistical inefficiency <math>g</math>. Bottom: Number of effectively uncorrelated samples <math>N_{eff}</math>. | 1 |
| 15:44, 22 December 2013 | Hamiltonian-exchange-reduced-potential.png (file) |  |
51 KB | John Chodera | Reduced potential as a function of iteration for Hamiltonian exchange simulation, computed as : <math> u(X_t) = \sum_{k=1}^K u_{s_{kt}}(x_{kt}) </math> where <math>u_i(x)</math> is the reduced potential for thermodynamic state index <math>i</math> and ... | 1 |
| 17:33, 26 September 2013 | Expanded.mdp (file) | 6 KB | Michael Shirts | Made the single precision soft core parameters the default. | 7 | |
| 17:29, 26 September 2013 | Ethanol.mdp (file) | 5 KB | Michael Shirts | Changed soft core parameters to default to good ones for single precision. | 3 | |
| 12:14, 27 July 2013 | FKBP AMBER GAFF.tgz (file) | 5.29 MB | Michael Shirts | Input files for ligands LG2,LG3, LG5, LG8, LG9, L12, L14, L20 bound to FKBP. | 1 | |
| 07:43, 13 July 2013 | Ligandtranslate.png (file) |  |
20 KB | Michael Shirts | 2 | |
| 17:28, 11 July 2013 | Samplecode.png (file) |  |
210 KB | Lnaden | Quick image for sample code used in the How-to box | 1 |
| 16:11, 3 January 2013 | Ethanol.top (file) | 4 KB | Michael Shirts | Gromacs .top file for Ethanol in TIP3P water parameterized with OPLS-AA | 1 | |
| 16:09, 3 January 2013 | Ethanol.gro (file) | 182 KB | Michael Shirts | A gro file for ethanol in water. | 1 | |
| 10:57, 26 October 2012 | T4-Tol-Imp-Well.png (file) | 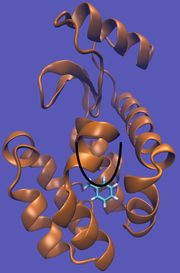 |
135 KB | Lnaden | Added a well to the T4-Toluene system | 1 |
| 10:36, 26 October 2012 | T4-Tol-Pull.gif (file) |  |
713 KB | Lnaden | Pulling animation for toluene | 1 |
| 14:20, 25 October 2012 | Implicit color.png (file) |  |
880 bytes | Lnaden | Empty Implicit Solvent Box | 1 |
| 14:15, 25 October 2012 | T4-Imp.png (file) |  |
130 KB | Lnaden | T4 in implicit solvent without toluene present | 1 |
| 14:14, 25 October 2012 | T4-Tol-Imp.png (file) |  |
134 KB | Lnaden | T4 Lysozyme and Toluene in an implicit solvent | 1 |
| 14:13, 25 October 2012 | Tol-Nosol.png (file) |  |
40 KB | Lnaden | Toluene up close, again but for tandem use with example | 1 |
| 14:12, 25 October 2012 | Tol-Imp.png (file) |  |
47 KB | Lnaden | Implicit solvent toluene | 1 |
| 16:47, 16 October 2012 | Ethanol in vac small.png (file) |  |
3 KB | Lnaden | Ethanol in a vacuum environment, small picture to better see whats going on. Rendered in VMD | 1 |
| 16:46, 16 October 2012 | TIP3P-Ethanol small.png (file) |  |
17 KB | Lnaden | Small water box with an ethanol in place. Not equilibrated, rendered in VMD | 1 |
| 16:45, 16 October 2012 | TIP3PBox small.png (file) |  |
13 KB | Lnaden | small TIP3PBox with an ethanol cavity. Not equilibrated, rendered in VMD | 1 |
| 16:37, 10 October 2012 | Toluene Phenol Topology.png (file) |  |
73 KB | Lnaden | 1 | |
| 13:54, 10 October 2012 | Doublearrowupdown.png (file) |  |
1 KB | Lnaden | Double arrow, pointing down and up | 1 |
| 09:54, 10 October 2012 | Blackbox.png (file) |  |
1 KB | Lnaden | A nice simple black box, for use in examples | 1 |
| 09:17, 10 October 2012 | Plus sign.jpg (file) |  |
2 KB | Lnaden | Simple plus sign, for use in examples | 1 |
| 09:12, 10 October 2012 | Ethanol in vac.png (file) |  |
4 KB | Lnaden | Ethanol by itself, but the boundary is expanded to show a "box" that it is free to explore. All atom, rendered in VMD | 1 |
| 08:57, 10 October 2012 | Waterbox.png (file) |  |
174 KB | Lnaden | New water box based on where an ethanol should be instead of ethane. | 2 |
| 08:55, 10 October 2012 | TIP3P-Ethanol.png (file) |  |
178 KB | Lnaden | Ethanol in a TIP3P water box. All atom, rendered in VMD | 1 |
| 08:55, 10 October 2012 | Ethanol.png (file) |  |
23 KB | Lnaden | Ethanol, all atom, rendered in VMD | 1 |
| 14:53, 9 October 2012 | Doublearrow.png (file) |  |
1 KB | Lnaden | 1 | |
| 13:41, 9 October 2012 | Toluene.png (file) |  |
44 KB | Lnaden | Toluene, Rendered in VMD | 1 |
| 13:40, 9 October 2012 | T4-Toluene.png (file) |  |
86 KB | Lnaden | T4-lysozyme with a Toluene at the binding site, Rendered in VMD | 1 |
| 13:40, 9 October 2012 | T4-Phenol.png (file) |  |
89 KB | Lnaden | T4-lysozyme with a Phenol at the binding site, Rendered in VMD | 1 |
| 13:39, 9 October 2012 | T4-lysozyme.png (file) |  |
83 KB | Lnaden | T4-lysozyme protein in implicit solvent. Rendered in VMD | 1 |